Welcome to deepCINAC’s documentation!¶
Calcium imaging toolbox¶
We have developed a Graphical User Interface (GUI) that offers various tools to visually evaluate inferred neuronal activity from inference methods and to build a eye inspection-based ground truth on calcium imaging data.
Then, we have designed a deep-learning based method, named DeepCINAC (Calcium Imaging Neuronal Activity Classifier). Instead of basing activity inference on the extracted fluorescence signal, DeepCINAC builds up on the visual inspection of each cell from the raw movie using the GUI.
This toolbox being very flexible, it can be adapted to any kind of calcium imaging dataset, in-vivo or in-vitro.
The toolbox also allows to predict cell type.
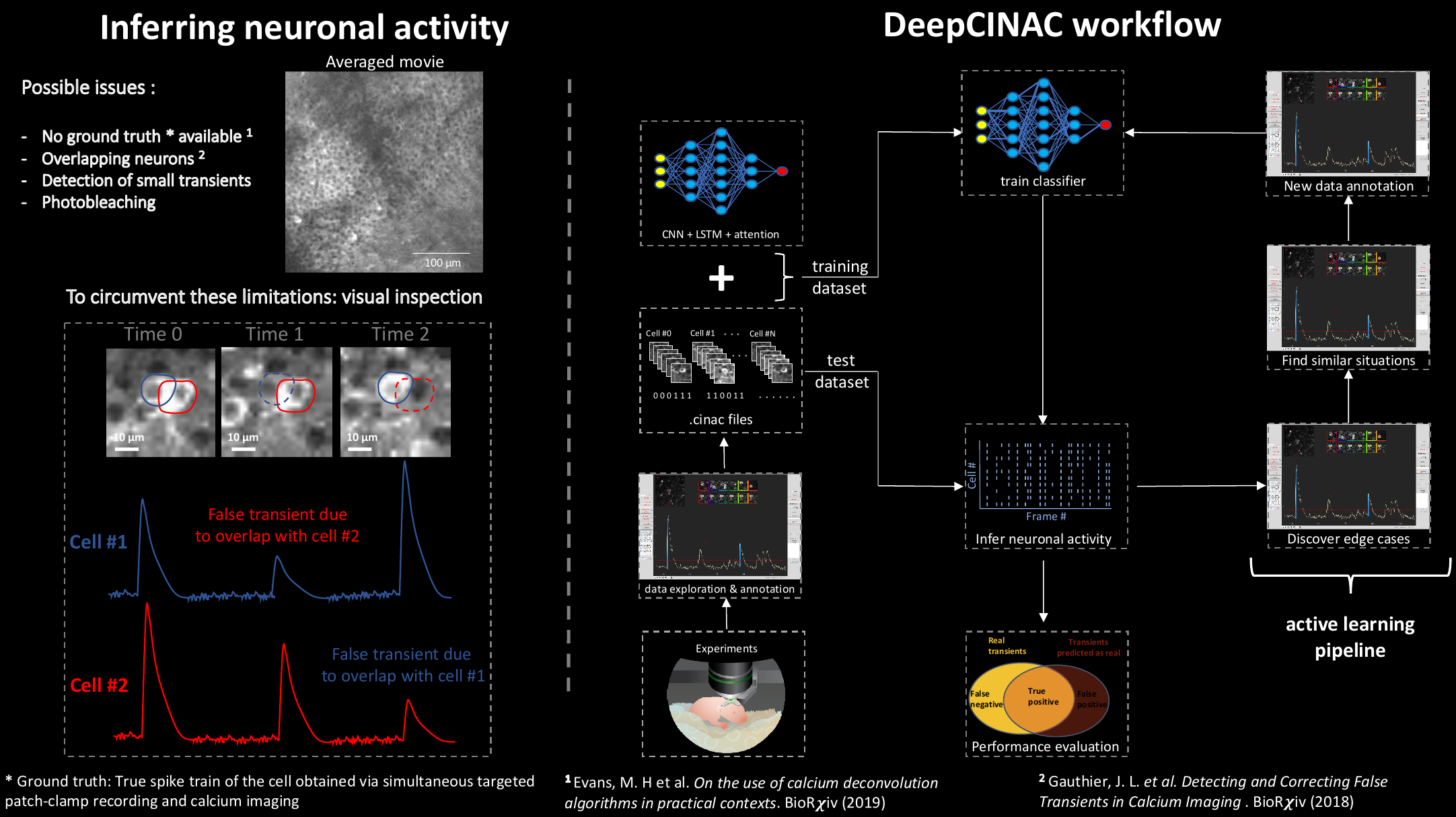
To train a classifier, the first step is to annotate data using the GUI. See the GUI tutorial for more information.
Then using the .cinac files produced, follow the instructions to train your classifier.
Note that .cinac files don’t contain the full original calcium imaging movie, only patches surrounding the cells you have annotated, so you can share the files without fearing that your data will be used by someone else other than to train a classifier.
To predict data, you can either use one of our pre-trained classifier or one you have trained, follow those instructions here .